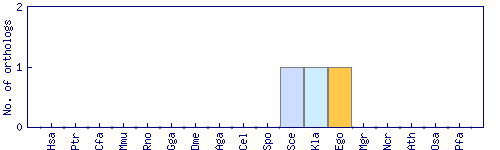
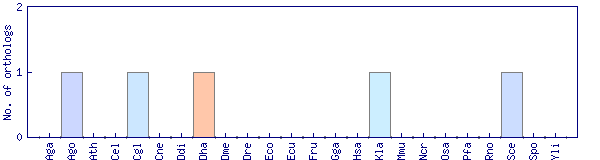
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | GSM1 | ||||||||||
| SGD link | S000003639 | ||||||||||
| Alternative ID | YJL103C | ||||||||||
| Description | Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis | ||||||||||
| Synonyms | YJL103C | ||||||||||
| Datasets |
|