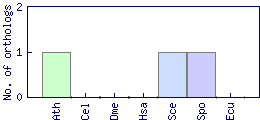
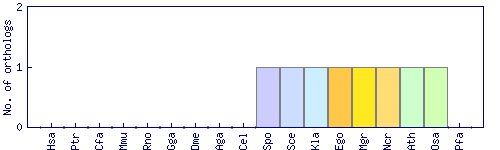
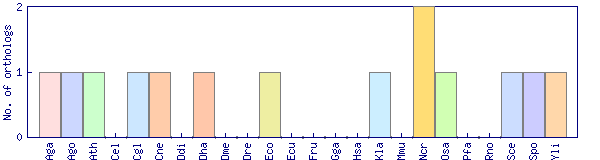
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ILV3 | ||||||||||
| SGD link | S000003777 | ||||||||||
| Alternative ID | YJR016C | ||||||||||
| Description | Dihydroxyacid dehydratase, catalyzes third step in the common pathway leading to biosynthesis of branched-chain amino acids | ||||||||||
| Synonyms | YJR016C | ||||||||||
| Datasets |
|