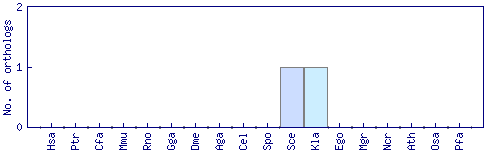
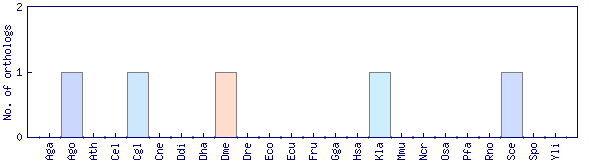
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | POL32 | ||||||||||
| SGD link | S000003804 | ||||||||||
| Alternative ID | YJR043C | ||||||||||
| Description | Third subunit of DNA polymerase delta, involved in chromosomal DNA replication; required for error-prone DNA synthesis in the presence of DNA damage and processivity; interacts with Hys2p, PCNA (Pol30p), and Pol1p | ||||||||||
| Synonyms | YJR043C | ||||||||||
| Datasets |
|