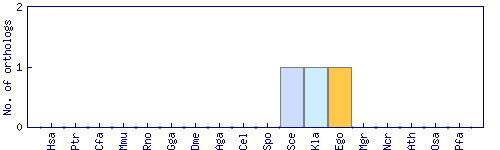
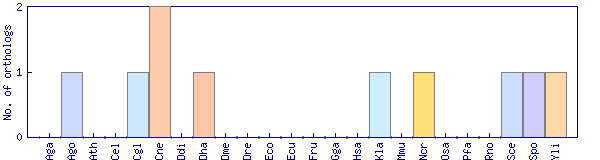
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | NTA1 | ||||||||||
| SGD link | S000003823 | ||||||||||
| Alternative ID | YJR062C | ||||||||||
| Description | Amidase, removes the amide group from N-terminal asparagine and glutamine residues to generate proteins with N-terminal aspartate and glutamate residues that are targets of ubiquitin-mediated degradation | ||||||||||
| Synonyms | YJR062C | ||||||||||
| Datasets |
|