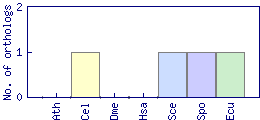
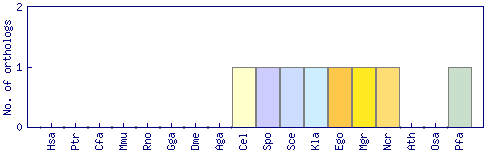
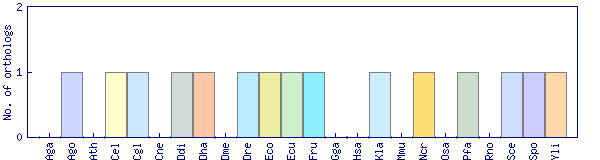
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | APN1 | ||||||||||
| SGD link | S000001597 | ||||||||||
| Alternative ID | YKL114C | ||||||||||
| Description | Major apurinic/apyrimidinic endonuclease, 3'-repair diesterase involved in repair of DNA damage by oxidation and alkylating agents; also functions as a 3'-5' exonuclease to repair 7,8-dihydro-8-oxodeoxyguanosine | ||||||||||
| Synonyms | YKL114C | ||||||||||
| Datasets |
|