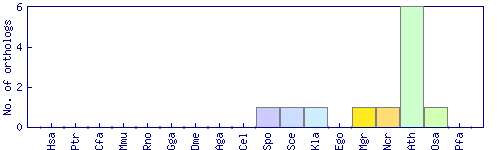
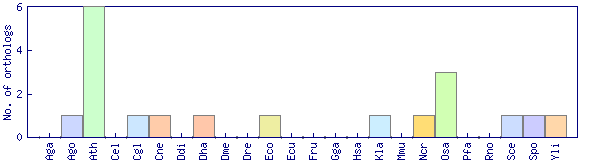
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | TRP3 | ||||||||||
| SGD link | S000001694 | ||||||||||
| Alternative ID | YKL211C | ||||||||||
| Description | Bifunctional enzyme exhibiting both indole-3-glycerol-phosphate synthase and anthranilate synthase activities, forms multifunctional hetero-oligomeric anthranilate synthase:indole-3-glycerol phosphate synthase enzyme complex with Trp2p | ||||||||||
| Synonyms | YKL211C | ||||||||||
| Datasets |
|