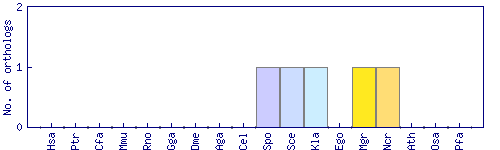
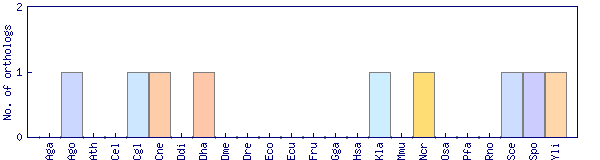
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | RAX2 | ||||||||||
| SGD link | S000004074 | ||||||||||
| Alternative ID | YLR084C | ||||||||||
| Description | N-glycosylated protein involved in the maintenance of bud site selection during bipolar budding; localization requires Rax1p; RAX2 mRNA stability is regulated by Mpt5p | ||||||||||
| Synonyms | YLR084C | ||||||||||
| Datasets |
|