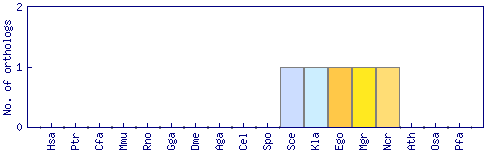
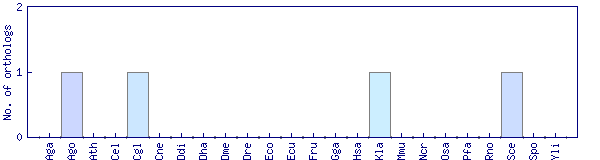
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | CHA4 | ||||||||||
| SGD link | S000004088 | ||||||||||
| Alternative ID | YLR098C | ||||||||||
| Description | DNA binding transcriptional activator, mediates serine/threonine activation of the catabolic L-serine (L-threonine) deaminase (CHA1); Zinc-finger protein with Zn[2]-Cys[6] fungal-type binuclear cluster domain | ||||||||||
| Synonyms | YLR098C | ||||||||||
| Datasets |
|