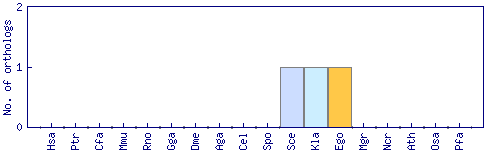
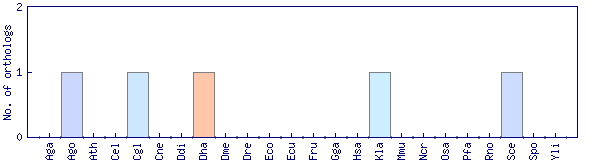
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | SLX4 | ||||||||||
| SGD link | S000004125 | ||||||||||
| Alternative ID | YLR135W | ||||||||||
| Description | Endonuclease involved in processing DNA during recombination and repair; cleaves branched structures in a complex with Slx1p; involved in Rad1p/Rad10p-dependent removal of 3'-nonhomologous tails during DSBR via single-strand annealing | ||||||||||
| Synonyms | YLR135W | ||||||||||
| Datasets |
|