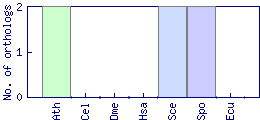
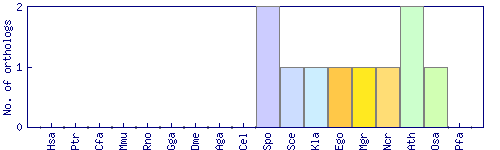
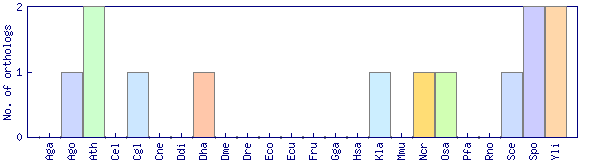
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ACF2 | ||||||||||
| SGD link | S000004134 | ||||||||||
| Alternative ID | YLR144C | ||||||||||
| Description | Intracellular beta-1,3-endoglucanase, expression is induced during sporulation; may have a role in cortical actin cytoskeleton assembly | ||||||||||
| Synonyms | YLR144C | ||||||||||
| Datasets |
|