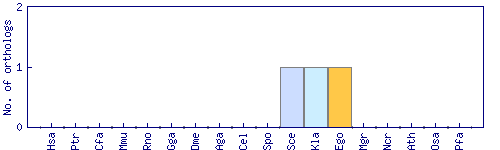
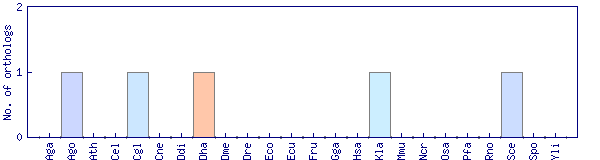
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | MEC3 | ||||||||||
| SGD link | S000004279 | ||||||||||
| Alternative ID | YLR288C | ||||||||||
| Description | DNA damage and meiotic pachytene checkpoint protein; subunit of a heterotrimeric complex (Rad17p-Mec3p-Ddc1p) that forms a sliding clamp, loaded onto partial duplex DNA by a clamp loader complex; homolog of human and S. pombe Hus1 | ||||||||||
| Synonyms | YLR288C | ||||||||||
| Datasets |
|