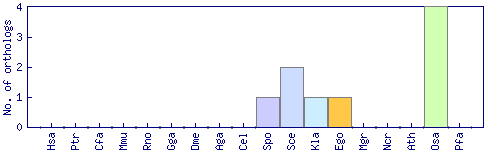
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | EXG1 | ||||||||||
| SGD link | S000004291 | ||||||||||
| Alternative ID | YLR300W | ||||||||||
| Description | Major exo-1,3-beta-glucanase of the cell wall, involved in cell wall beta-glucan assembly; exists as three differentially glycosylated isoenzymes | ||||||||||
| Synonyms | YLR300W | ||||||||||
| Datasets |
|