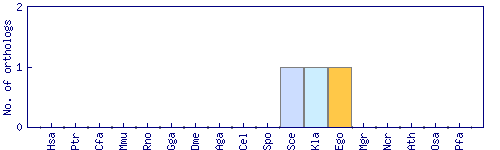
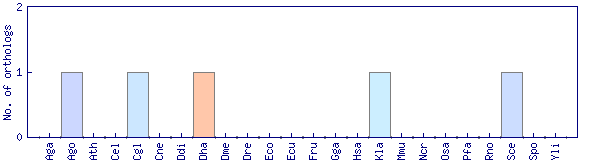
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | MMS22 | ||||||||||
| SGD link | S000004312 | ||||||||||
| Alternative ID | YLR320W | ||||||||||
| Description | Protein that acts with Mms1p in a repair pathway that may be involved in resolving replication intermediates or preventing the damage caused by blocked replication forks; required for accurate meiotic chromosome segregation | ||||||||||
| Synonyms | YLR320W | ||||||||||
| Datasets |
|