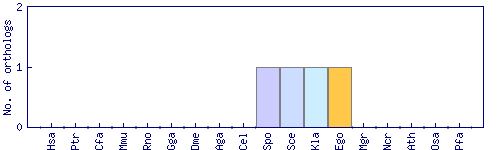
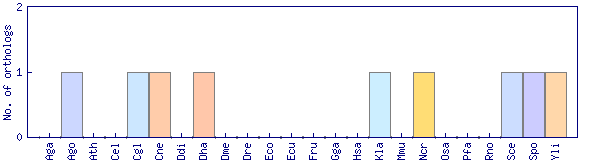
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | NUP2 | ||||||||||
| SGD link | S000004327 | ||||||||||
| Alternative ID | YLR335W | ||||||||||
| Description | Nucleoporin involved in nucleocytoplasmic transport, binds to either the nucleoplasmic or cytoplasmic faces of the nuclear pore complex depending on Ran-GTP levels; also has a role in chromatin organization | ||||||||||
| Synonyms | YLR335W | ||||||||||
| Datasets |
|