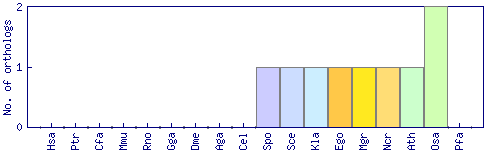
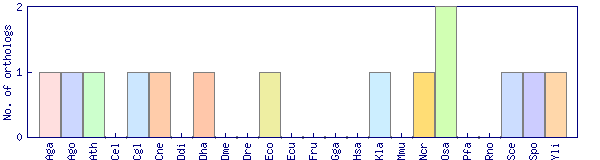
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ILV5 | ||||||||||
| SGD link | S000004347 | ||||||||||
| Alternative ID | YLR355C | ||||||||||
| Description | Acetohydroxyacid reductoisomerase, mitochondrial protein involved in branched-chain amino acid biosynthesis, also required for maintenance of wild-type mitochondrial DNA and found in mitochondrial nucleoids | ||||||||||
| Synonyms | YLR355C | ||||||||||
| Datasets |
|