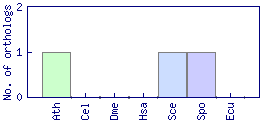
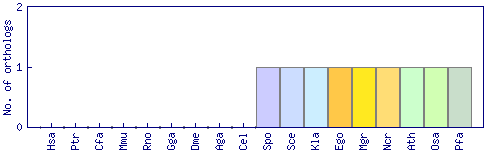
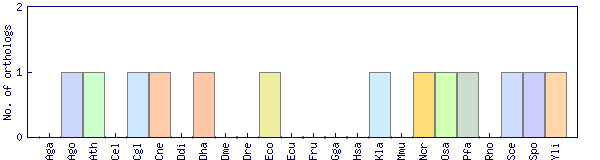
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | URA4 | ||||||||||
| SGD link | S000004412 | ||||||||||
| Alternative ID | YLR420W | ||||||||||
| Description | Dihydroorotase, catalyzes the third enzymatic step in the de novo biosynthesis of pyrimidines, converting carbamoyl-L-aspartate into dihydroorotate | ||||||||||
| Synonyms | YLR420W | ||||||||||
| Datasets |
|