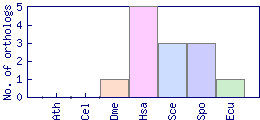
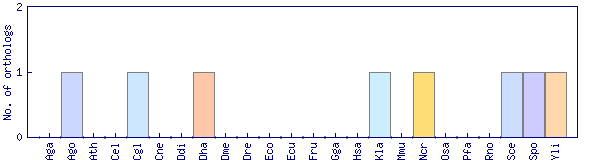
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | TUS1 | ||||||||||
| SGD link | S000004417 | ||||||||||
| Alternative ID | YLR425W | ||||||||||
| Description | Guanine nucleotide exchange factor (GEF) that functions to modulate Rho1p activity as part of the cell integrity signaling pathway; multicopy suppressor of tor2 mutation and ypk1 ypk2 double mutation; potential Cdc28p substrate | ||||||||||
| Synonyms | YLR425W | ||||||||||
| Datasets |
|