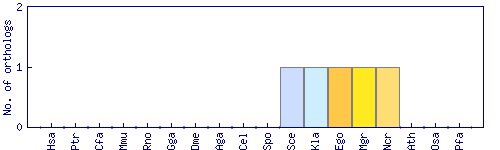
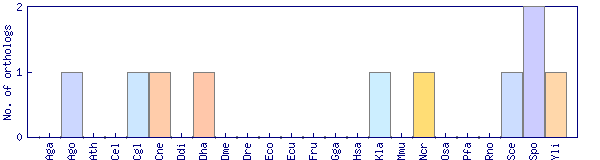
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | SRC1 | ||||||||||
| SGD link | S000004497 | ||||||||||
| Alternative ID | YML034W | ||||||||||
| Description | Inner nuclear membrane protein that functions in regulation of subtelomeric genes and is linked to TREX (transcription export) factors; SRC1 produces 2 splice variant proteins with different functions; possible role in chromatid segregation | ||||||||||
| Synonyms | YML034W | ||||||||||
| Datasets |
|