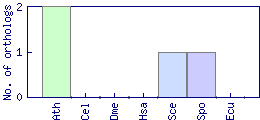
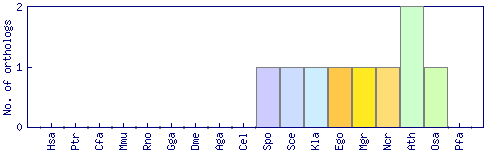
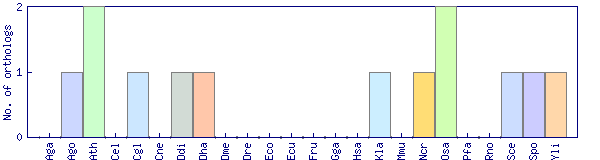
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | TEM1 | ||||||||||
| SGD link | S000004529 | ||||||||||
| Alternative ID | YML064C | ||||||||||
| Description | GTP-binding protein of the ras superfamily involved in termination of M-phase; controls actomyosin and septin dynamics during cytokinesis | ||||||||||
| Synonyms | YML064C | ||||||||||
| Datasets |
|