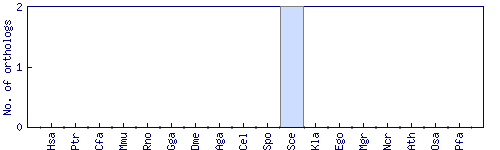
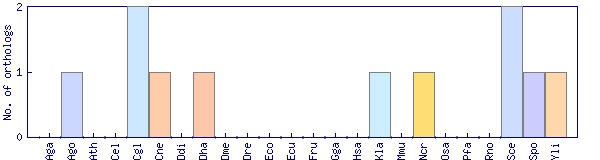
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ZDS2 | ||||||||||
| SGD link | S000004577 | ||||||||||
| Alternative ID | YML109W | ||||||||||
| Description | Protein that interacts with silencing proteins at the telomere, involved in transcriptional silencing; implicated in the mitotic exit network through regulation of Cdc14p localization; paralog of Zds1p | ||||||||||
| Synonyms | YML109W | ||||||||||
| Datasets |
|