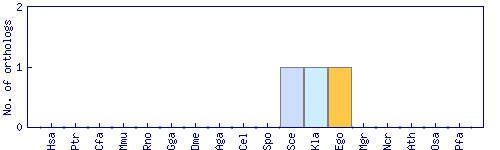
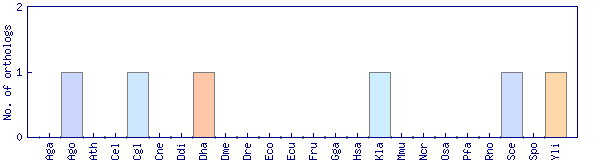
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | VAN1 | ||||||||||
| SGD link | S000004583 | ||||||||||
| Alternative ID | YML115C | ||||||||||
| Description | Component of the mannan polymerase I, which contains Van1p and Mnn9p and is involved in the first steps of mannan synthesis; mutants are vanadate-resistant | ||||||||||
| Synonyms | YML115C | ||||||||||
| Datasets |
|