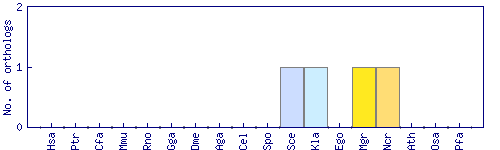
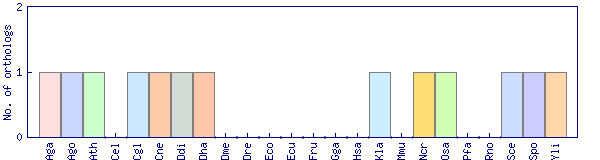
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | CCS1 | ||||||||||
| SGD link | S000004641 | ||||||||||
| Alternative ID | YMR038C | ||||||||||
| Description | Copper chaperone for superoxide dismutase Sod1p, involved in oxidative stress protection; Met-X-Cys-X2-Cys motif within the N-terminal portion is involved in insertion of copper into Sod1p under conditions of copper deprivation | ||||||||||
| Synonyms | YMR038C | ||||||||||
| Datasets |
|