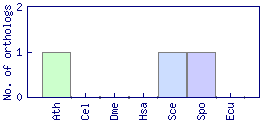
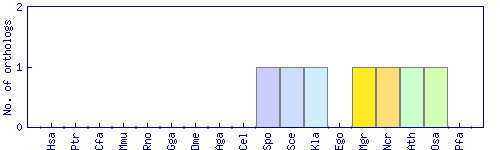
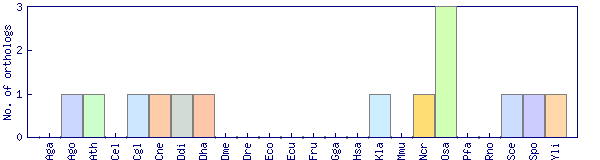
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ARG7 | ||||||||||
| SGD link | S000004666 | ||||||||||
| Alternative ID | YMR062C | ||||||||||
| Description | Mitochondrial ornithine acetyltransferase, catalyzes the fifth step in arginine biosynthesis; also possesses acetylglutamate synthase activity, regenerates acetylglutamate while forming ornithine | ||||||||||
| Synonyms | YMR062C | ||||||||||
| Datasets |
|