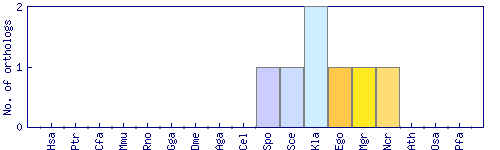
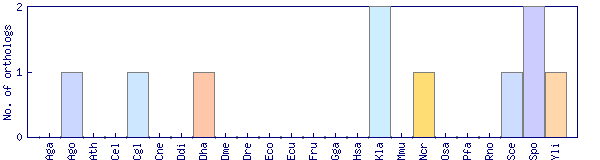
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | RIM9 | ||||||||||
| SGD link | S000004667 | ||||||||||
| Alternative ID | YMR063W | ||||||||||
| Description | Protein of unknown function, involved in the proteolytic activation of Rim101p in response to alkaline pH; has similarity to A. nidulans PalI; putative membrane protein | ||||||||||
| Synonyms | YMR063W | ||||||||||
| Datasets |
|