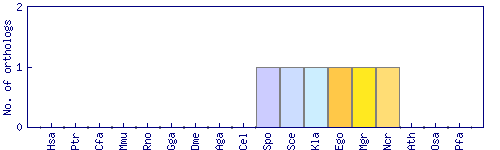
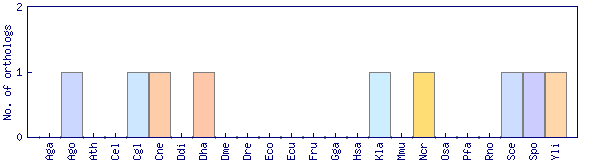
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | NPL6 | ||||||||||
| SGD link | S000004697 | ||||||||||
| Alternative ID | YMR091C | ||||||||||
| Description | Component of the RSC chromatin remodeling complex; interacts with Rsc3p, Rsc30p, Ldb7p, and Htl1p to form a module important for a broad range of RSC functions; involved in nuclear protein import and maintenance of proper telomere length | ||||||||||
| Synonyms | YMR091C | ||||||||||
| Datasets |
|