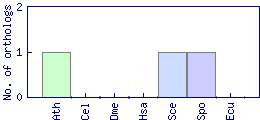
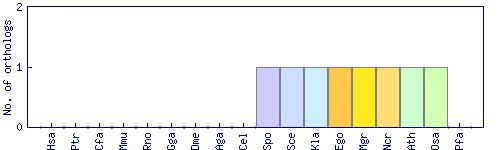
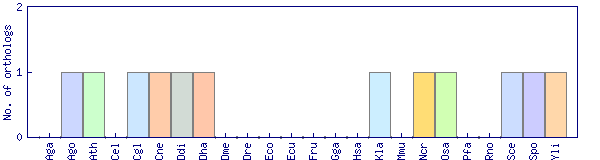
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ERG8 | ||||||||||
| SGD link | S000004833 | ||||||||||
| Alternative ID | YMR220W | ||||||||||
| Description | Phosphomevalonate kinase, an essential cytosolic enzyme that acts in the biosynthesis of isoprenoids and sterols, including ergosterol, from mevalonate | ||||||||||
| Synonyms | YMR220W | ||||||||||
| Datasets |
|