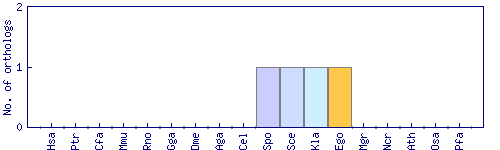
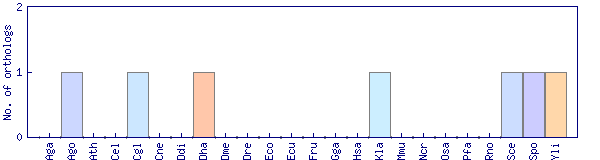
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | MTF1 | ||||||||||
| SGD link | S000004841 | ||||||||||
| Alternative ID | YMR228W | ||||||||||
| Description | Mitochondrial RNA polymerase specificity factor with structural similarity to S-adenosylmethionine-dependent methyltransferases and functional similarity to bacterial sigma-factors, interacts with mitochondrial core polymerase Rpo41p | ||||||||||
| Synonyms | YMR228W | ||||||||||
| Datasets |
|