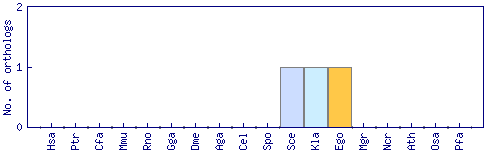
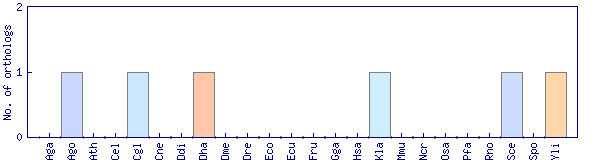
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | SAP30 | ||||||||||
| SGD link | S000004876 | ||||||||||
| Alternative ID | YMR263W | ||||||||||
| Description | Subunit of a histone deacetylase complex, along with Rpd3p and Sin3p, that is involved in silencing at telomeres, rDNA, and silent mating-type loci; involved in telomere maintenance | ||||||||||
| Synonyms | YMR263W | ||||||||||
| Datasets |
|