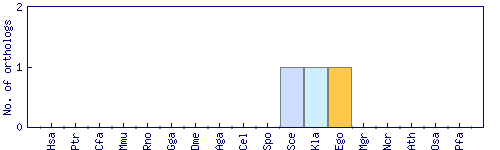
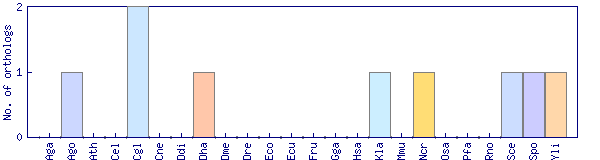
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | CUE1 | ||||||||||
| SGD link | S000004877 | ||||||||||
| Alternative ID | YMR264W | ||||||||||
| Description | Endoplasmic reticulum membrane protein that recruits the ubiquitin-conjugating enzyme Ubc7p to the ER where it functions in protein degradation; contains a CUE domain that binds ubiquitin to facilitate intramolecular monoubiquitination | ||||||||||
| Synonyms | YMR264W | ||||||||||
| Datasets |
|