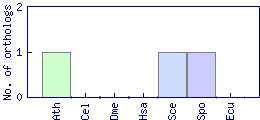
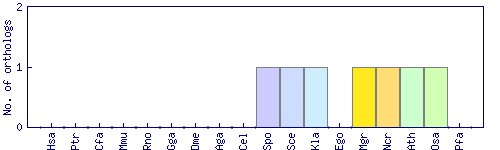
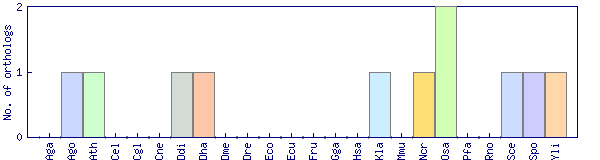
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | RIT1 | ||||||||||
| SGD link | S000004896 | ||||||||||
| Alternative ID | YMR283C | ||||||||||
| Description | 2'-O-ribosyl phosphate transferase, modifies the initiator methionine tRNA at position 64 to distinguish it from elongator methionine tRNA | ||||||||||
| Synonyms | YMR283C | ||||||||||
| Datasets |
|