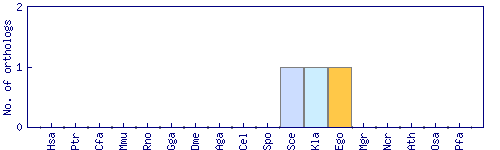
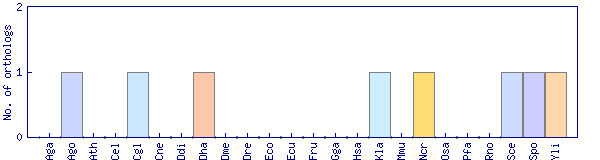
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ABZ2 | ||||||||||
| SGD link | S000004902 | ||||||||||
| Alternative ID | YMR289W | ||||||||||
| Description | Aminodeoxychorismate lyase (4-amino-4-deoxychorismate lyase), catalyzes the third step in para-aminobenzoic acid biosynthesis; involved in folic acid biosynthesis | ||||||||||
| Synonyms | YMR289W | ||||||||||
| Datasets |
|