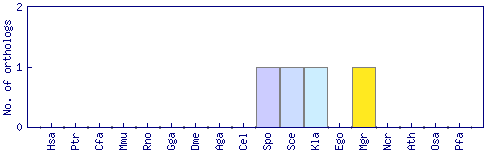
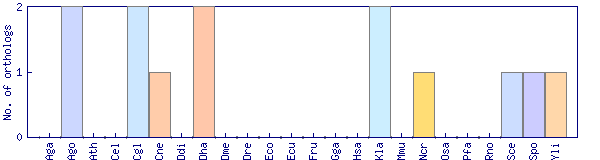
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | GAS1 | ||||||||||
| SGD link | S000004924 | ||||||||||
| Alternative ID | YMR307W | ||||||||||
| Description | Beta-1,3-glucanosyltransferase, required for cell wall assembly and also has a role in transcriptional silencing; localizes to the cell surface via a glycosylphosphatidylinositol (GPI) anchor; also found at the nuclear periphery | ||||||||||
| Synonyms | YMR307W | ||||||||||
| Datasets |
|