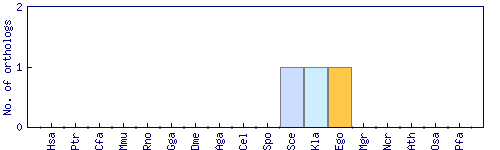
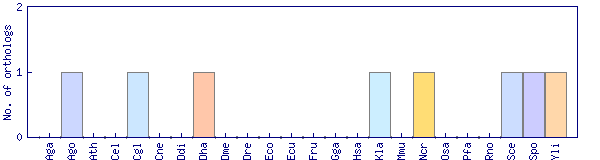
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ELP6 | ||||||||||
| SGD link | S000004929 | ||||||||||
| Alternative ID | YMR312W | ||||||||||
| Description | Subunit of Elongator complex, which is required for modification of wobble nucleosides in tRNA; required for Elongator structural integrity | ||||||||||
| Synonyms | YMR312W | ||||||||||
| Datasets |
|