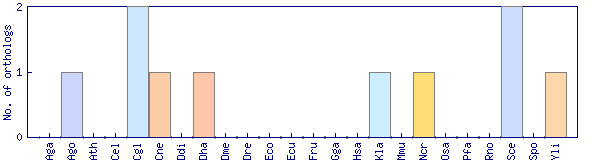
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ASI3 | ||||||||||
| SGD link | S000004953 | ||||||||||
| Alternative ID | YNL008C | ||||||||||
| Description | Putative integral membrane E3 ubiquitin ligase; acts with Asi1p and Asi2p to ensure the fidelity of SPS-sensor signalling by maintaining the dormant repressed state of gene expression in the absence of inducing signals | ||||||||||
| Synonyms | YNL008C | ||||||||||
| Datasets |
|