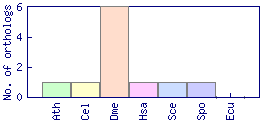
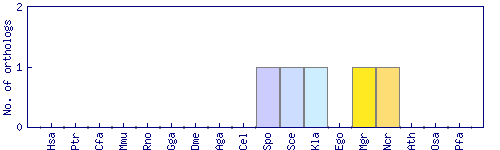
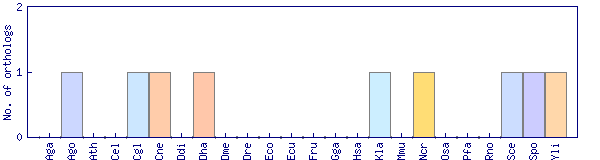
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | TOF1 | ||||||||||
| SGD link | S000005217 | ||||||||||
| Alternative ID | YNL273W | ||||||||||
| Description | Subunit of a replication-pausing checkpoint complex (Tof1p-Mrc1p-Csm3p) that acts at the stalled replication fork to promote sister chromatid cohesion after DNA damage, facilitating gap repair of damaged DNA; interacts with the MCM helicase | ||||||||||
| Synonyms | YNL273W | ||||||||||
| Datasets |
|