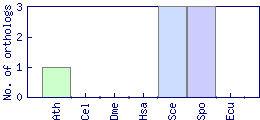
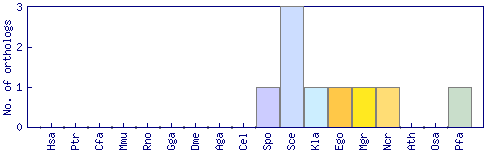
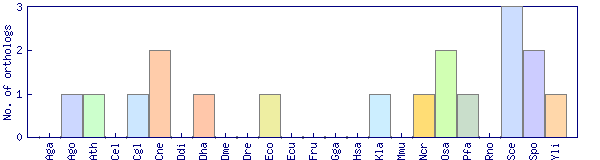
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | THI20 | ||||||||||
| SGD link | S000005416 | ||||||||||
| Alternative ID | YOL055C | ||||||||||
| Description | Multifunctional protein with hydroxymethylpyrimidine phosphate (HMP-P) kinase and thiaminase activities; involved in thiamine biosynthesis and degradation; in a gene family with THI21 and THI22; HMP-P kinase activity redundant with Thi21p | ||||||||||
| Synonyms | YOL055C | ||||||||||
| Datasets |
|