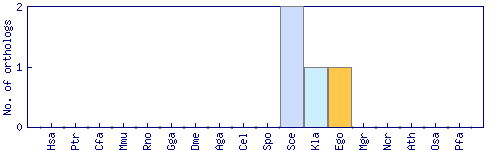
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | STD1 | ||||||||||
| SGD link | S000005573 | ||||||||||
| Alternative ID | YOR047C | ||||||||||
| Description | Protein involved in control of glucose-regulated gene expression; interacts with protein kinase Snf1p, glucose sensors Snf3p and Rgt2p, and TATA-binding protein Spt15p; acts as a regulator of the transcription factor Rgt1p | ||||||||||
| Synonyms | YOR047C | ||||||||||
| Datasets |
|