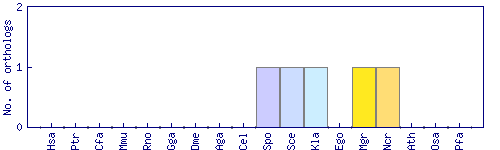
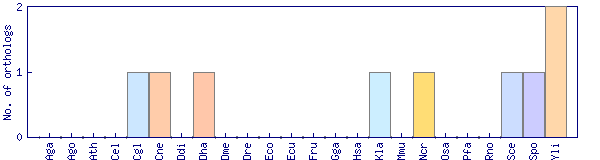
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | RSB1 | ||||||||||
| SGD link | S000005575 | ||||||||||
| Alternative ID | YOR049C | ||||||||||
| Description | Suppressor of sphingoid long chain base (LCB) sensitivity of an LCB-lyase mutation; putative integral membrane transporter or flippase that may transport LCBs from the cytoplasmic side toward the extracytoplasmic side of the membrane | ||||||||||
| Synonyms | YOR049C | ||||||||||
| Datasets |
|