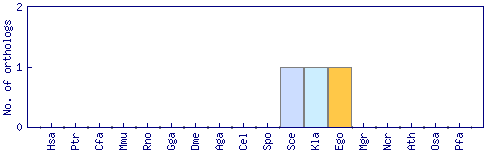
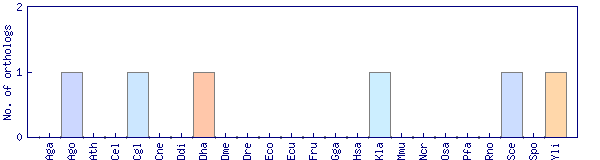
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | DIA2 | ||||||||||
| SGD link | S000005606 | ||||||||||
| Alternative ID | YOR080W | ||||||||||
| Description | Origin-binding F-box protein that forms an SCF ubiquitin ligase complex with Skp1p and Cdc53p; plays a role in DNA replication, involved in invasive and pseudohyphal growth | ||||||||||
| Synonyms | YOR080W | ||||||||||
| Datasets |
|