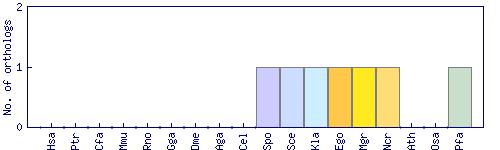
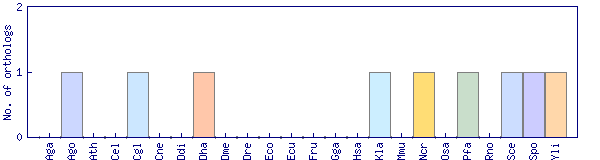
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ISN1 | ||||||||||
| SGD link | S000005681 | ||||||||||
| Alternative ID | YOR155C | ||||||||||
| Description | Inosine 5'-monophosphate (IMP)-specific 5'-nucleotidase, catalyzes the breakdown of IMP to inosine, does not show similarity to known 5'-nucleotidases from other organisms | ||||||||||
| Synonyms | YOR155C | ||||||||||
| Datasets |
|