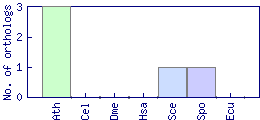
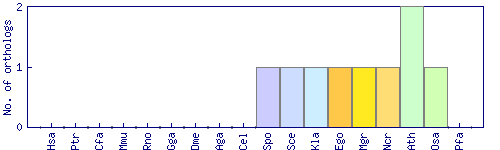
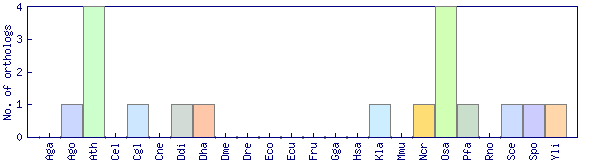
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | SEY1 | ||||||||||
| SGD link | S000005691 | ||||||||||
| Alternative ID | YOR165W | ||||||||||
| Description | GTPase with a role in ER morphology; interacts physically and genetically with Yop1p and Rtn1p; possible functional ortholog of mammalian atlastins, defects in which cause a form of hereditary spastic paraplegia; homolog of Arabidopsis RHD3 | ||||||||||
| Synonyms | YOR165W | ||||||||||
| Datasets |
|