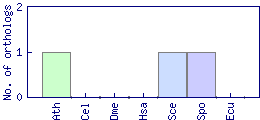
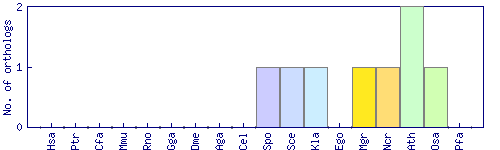
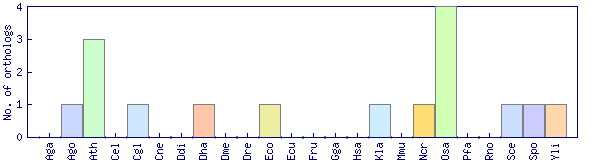
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | HIS3 | ||||||||||
| SGD link | S000005728 | ||||||||||
| Alternative ID | YOR202W | ||||||||||
| Description | Imidazoleglycerol-phosphate dehydratase, catalyzes the sixth step in histidine biosynthesis; mutations cause histidine auxotrophy and sensitivity to Cu, Co, and Ni salts; transcription is regulated by general amino acid control via Gcn4p | ||||||||||
| Synonyms | YOR202W | ||||||||||
| Datasets |
|