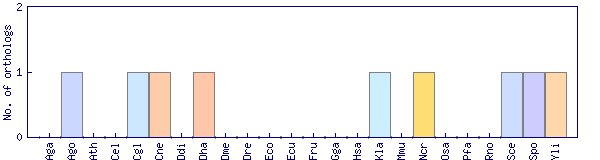
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | VTS1 | ||||||||||
| SGD link | S000005886 | ||||||||||
| Alternative ID | YOR359W | ||||||||||
| Description | Post-transcriptional gene regulator, flap-structured DNA-binding and RNA-binding protein; shows genetic interactions with Vti1p, a v-SNARE involved in cis-Golgi membrane traffic; stimulates Dna2p endonuclease activity; contains a SAM domain | ||||||||||
| Synonyms | YOR359W | ||||||||||
| Datasets |
|