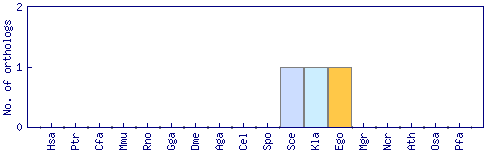
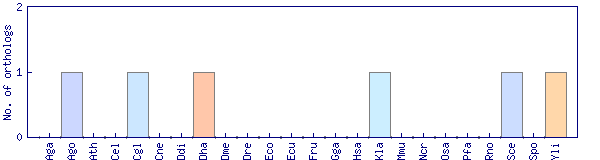
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | TCO89 | ||||||||||
| SGD link | S000006101 | ||||||||||
| Alternative ID | YPL180W | ||||||||||
| Description | Subunit of TORC1 (Tor1p or Tor2p-Kog1p-Lst8p-Tco89p), a complex that regulates growth in response to nutrient availability; cooperates with Ssd1p in the maintenance of cellular integrity; deletion strains are hypersensitive to rapamycin | ||||||||||
| Synonyms | YPL180W | ||||||||||
| Datasets |
|