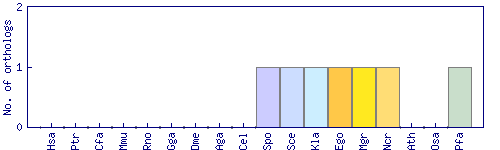
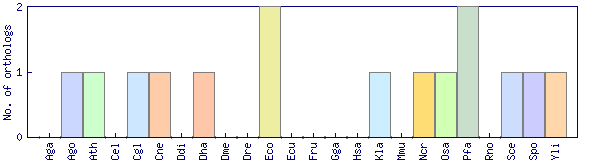
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | THI6 | ||||||||||
| SGD link | S000006135 | ||||||||||
| Alternative ID | YPL214C | ||||||||||
| Description | Bifunctional enzyme with thiamine-phosphate pyrophosphorylase and 4-methyl-5-beta-hydroxyethylthiazole kinase activities, required for thiamine biosynthesis; GFP-fusion protein localizes to the cytoplasm in a punctate pattern | ||||||||||
| Synonyms | YPL214C | ||||||||||
| Datasets |
|