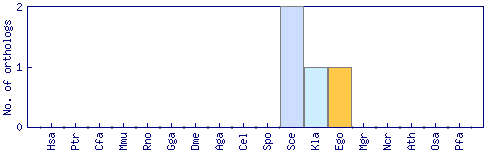
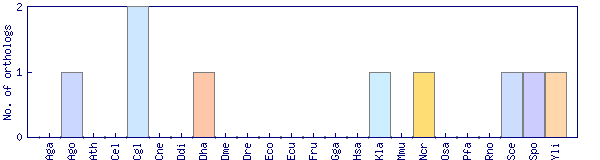
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | NCE102 | ||||||||||
| SGD link | S000006353 | ||||||||||
| Alternative ID | YPR149W | ||||||||||
| Description | Protein of unknown function; contains transmembrane domains; involved in secretion of proteins that lack classical secretory signal sequences; component of the detergent-insoluble glycolipid-enriched complexes (DIGs) | ||||||||||
| Synonyms | YPR149W | ||||||||||
| Datasets |
|