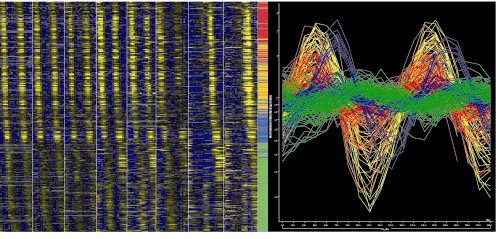
We report the genome-wide transcriptional program of the fission yeast cell cycle, identifying 407 periodically expressed genes, 136 of which exhibit high-amplitude changes. These genes cluster in four major waves of expression. The forkhead protein Sep1p regulates mitotic genes in the first cluster including Ace2p, which activates transcription in the second cluster during the M/G1 transition and cytokinesis. Other genes in the second cluster required for G1/S progression are regulated by the MBF-complex independently of Sep1p and Ace2p. The third cluster coincides with S-phase, whilst a fourth cluster contains genes weakly regulated during G2-phase. Below there are links to access various information relevant to our cell cycle data.
Rustici G, Mata J, Kivinen K, Lió P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, and Bähler J (2004).
Periodic gene expression program of the fission yeast cell cycle.
Nature Genet. 36:809-817.
A reprint is available as a PDF.
Gene expression viewer:
To check the expression value of your favourite gene use the search box below:
Use commas to separate multiple queries e.g. pat1,tea1.
Click on the gene name in the key (bottom of graph) to connect to GeneDB.
Supplementary Data:
- Supplementary Figure 1: Data on two synchronized timecourse experiments.
- Supplementary Table 1: Genes previously reported as cell cycle regulated.
- Supplementary Table 2: Periodic genes with characterized functions.
- Supplementary Table 3: List of 407 genes periodically expressed during the cell cycle.
- Supplementary Table 4: Enriched Gene Ontology (GO) terms among periodically expressed genes.
- Supplementary Table 5: Potential regulatory promoter motifs.
- Supplementary Table 6: Overlap of periodic genes between fission and budding yeasts.
- Supplementary Table 7: S. pombe strains used in this study.
Complete Normalized Data Sets:
-
Timecourse experiment of cell-cycle synchronized cells (elutriation 1): 20 timepoints,
2 cell cycles:
Download data (ftp) -
Timecourse experiment of cell-cycle synchronized cells (elutriation 2): 20 timepoints,
2 cell cycles:
Download data (ftp) -
Timecourse experiment of cell-cycle synchronized cells (elutriation 3): 20 timepoints,
2 cell cycles:
Download data (ftp) -
Timecourse experiment of cell-cycle synchronized cells (cdc25 block-release 1): 19 timepoints,
2 cell cycles:
Download data (ftp) -
Timecourse experiment of cell-cycle synchronized cells (cdc25 block-release 2): 18 timepoints,
technical replicates, 2 cell cycles:
Download data (ftp) -
Timecourse experiment of cell-cycle synchronized cells (cdc25 block-release in sep1 mutant background):
20 timepoints, 2 cell cycles:
Download data (ftp) -
Timecourse experiment of cell-cycle synchronized cells (elutriation and cdc10 block-release):
22 timepoints, G1 arrest and 1 cell cycle:
Download data (ftp) -
Timecourse experiment of cell-cycle synchronized cells (elutriation and cdc25 block-release):
21 timepoints, G2 arrest and 1 cell cycle:
Download data (ftp) -
Mutant experiment: sep1 mutant vs wild-type (4 biological repeats):
Download data (ftp) -
Mutant experiment: ace2 mutant vs wild-type (5 biological repeats):
Download data (ftp) -
Mutant experiment: cdc10-C4 mutant vs wild-type (4 biological repeats):
Download data (ftp) -
Overexpression experiment: Sep1p OE vs wild-type (2 biological repeats):
Download data (ftp) -
Overexpression experiment: Ace2p OE vs wild-type (2 biological repeats):
Download data (ftp) -
Replication block experiment: hydroxyurea block for 3 h vs unblocked cells:
Download data (ftp)
All raw data are available under accession numbers E-MEXP-54 to E-MEXP-64 from ArrayExpress.
