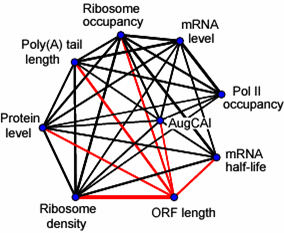
We applied a wide range of microarray-based approaches to determine key gene expression intermediates in exponentially growing fission yeast, providing genome-wide data for translational profiles, mRNA steady-state levels, polyadenylation profiles, start-codon sequence context, mRNA half- lives, and RNA polymerase II occupancy. These rich data sets, all acquired under a standardized condition, reveal a substantial coordination between regulatory layers and provide a basis for a systems-level understanding of multi-layered gene expression programs. Below there are links to access various information relevant to these data.
Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, and Bähler J (2007).
A Network of Multiple Regulatory Layers Shapes Gene Expression in Fission Yeast.
Mol. Cell 26:145-155.
A reprint is available as a PDF.
Supplementary Data:
- Supplementary Information: Supplemental Experimental Procedures, Figures, and References.
- Supplementary Table S1: Processed genome-wide data sets on ribosome occupancy, ribosome number, ribosome density, ORF length, poly(A) tail rank, mRNA level, relative Pol II occupancy, AugCAI, and mRNA half-life.
Complete Normalized Data Sets:
-
Translation profiling data (3 independent repeats of 12 fractions each):
Download data (ftp) -
Polyadenylation profiling data/PASTA analysis (2 independent repeats of 5 fractions each):
Download data (ftp) -
Steady-state mRNA level data (2 independent Affymetrix experiments):
Download data (ftp) -
mRNA decay data (3 independent timecourses of 3 timepoints each):
Download data (ftp) -
Polymerase II occupancy data/transcriptional efficiency (2 independent repeats):
Download data (ftp)
All raw data are available under accession numbers E-TABM-217 to E-TABM-220
and E-TABM-222 from ArrayExpress.