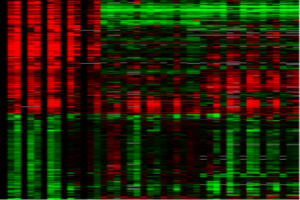
Cells have to adjust to changes in their environment and launch defence mechanisms against potentially life threatening conditions. We are studying global stress responses and their regulation using various environmental stresses (oxidative stress, heavy metal stress, osmotic stress, heat shock, and DNA damage). Special emphasis is on oxidative stress, where we are comparing responses to different doses and oxidants as well as analysing underlying regulatory mechanisms. This work is performed in collaboration with Nic Jones' Group (Paterson Institute, Manchester, UK).
Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma
A, Jones N, and Bähler J (2003). Global transcriptional responses of
fission yeast to environmental stress.
Mol. Biol.
Cell 14:214-229.
A reprint is available as a PDF.
Gene expression viewer:
To check the expression value of your favourite gene use the search box below:
Use commas to separate multiple queries e.g. pat1,tea1.
Click on the gene name in the key (bottom of graph) to connect to GeneDB.
Gene Lists (as defined in above paper)
- Stress gene look-up table: (genes induced >3-fold in at least one stress and their dependency on sty1 and/or atf1).
- Induced CESR genes: (140 genes, conservative list using cutoffs, Table 1).
- Induced CESR genes: (314 genes, less conservative list using clustering).
- Repressed CESR genes: (106 genes, conservative list using cutoffs).
- Repressed CESR genes: (424 genes, less conservative list using clustering).
- Stress genes induced in budding and fission yeasts: (49 genes, Figure 3).
- Stress genes repressed in budding and fission yeasts: (221 genes, Figure 3).
- Hydrogen peroxide stress genes: (56 specific and 12 super-induced genes).
- Cadmium stress genes: (32 specific and 2 super-induced genes).
- Heat shock genes: (20 specific and 11 super-induced genes).
- Osmotic stress genes: (13 specific and 8 super-induced genes).
- Clusters 1-7: (Figure 5A).
- Unstressed cells: (genes dependent on sty1- and/or atf1 in absence of stress).
- Promoter motifs: (genes dependent on sty1- and/or atf1 during stress; Table 2).
Complete Data Sets
-
Five different stress timecourse experiments with wild type cells: average of two independent experiments each:
Download data (ftp) -
Five different stress timecourse experiments with wild type cells: separate results for two independent experiments:
Download data (ftp) -
Five different stress timecourse experiments with sty1Δ mutants: average of two independent experiments each:
Download data (ftp) -
Five different stress timecourse experiments with sty1Δ mutants: separate results for two independent experiments:
Download data (ftp) -
Five different stress timecourse experiments with atf1Δ mutants: single experiment each:
Download data (ftp)
All raw data are available from accession number E-MEXP-29 in
ArrayExpress.
Multiple pathways differentially regulate global oxidative stress responses
Chen D, Wilkinson CR, Watt S, Penkett CJ, Toone WM, Jones N, and Bähler J (2008).Multiple pathways differentially regulate global oxidative stress responses in fission yeast.
Mol. Biol.
Cell 19:308-317.
A reprint is available as a PDF.
Supplementary material:
- Supplemental Figure 1 (pdf).
- Supplemental Table 1 and 2 (pdf).
- Supplemental Table 3 (xls).
Complete Data Sets:
Figure numbers refer to the corresponding figures in the paper, defining different combinations of conditions.
- Figure 1A Download data.
- Figure 2 Download Data.
- Figure 3 Download Data.
- Figure 4 Download Data.
- Figure 5 Download Data.
- Figure 7B Download Data.
All raw data are available from accession number E-MEXP-1083 in
ArrayExpress.
